REGINDEX
Compressed Indexes for Regular Languages with Applications to Computational Pan-genomics
Project
Sorting is, arguably, the most powerful algorithmic building block when it comes to indexing data. At the same time, the regularities exposed by sorting are precisely those enabling data compression. In the last two decades, this fascinating duality has led researchers to the design of compressed full-text indexes: data structures supporting fast pattern matching queries over compressed text. In this project, we revisit the natural generalization of the problem to labeled graphs from a new perspective: we interpret graphs as finite-state automata and investigate the connections existing between their propensity to be sorted and the regular languages they recognize. This novel language-theoretic approach makes it possible to transfer fundamental results between the mature fields of regular language theory and compressed text indexing. The project aims at building this bridge by developing a new theory of compressed regular language indexing.
The project finds important applications to the rapidly-expanding field of computational pangenomics, where the goal is to study the variations contained in the genomes of an entire population. Recent research has shown that representing pan-genomes as labeled graphs is an important step to reduce reference allele bias. Existing approaches, however, can index only restricted classes of graphs, thereby limiting the practical applicability of such powerful pan-genome representations.
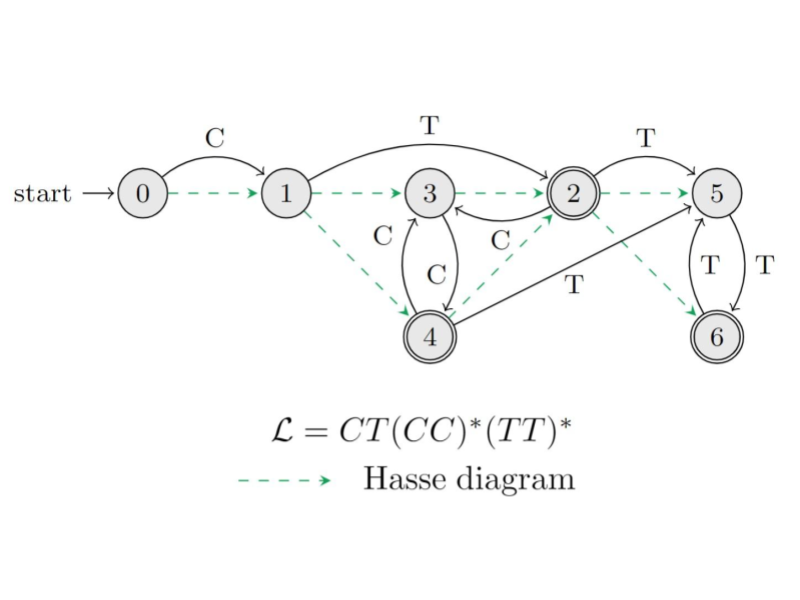
The project’s approach, based on sorting regular languages by partial co-lexicographic orders (see figure), changes the perspective from which the compressed indexing problem has been tackled in the literature. This project aims at developing a theory of graph indexing and compression based on the natural interplay between sorting and regular language theory.
Publications
Journal papers
- Elena Biagi, Davide Cenzato, Zsuzsanna Lipták, Giuseppe Romana. On the number of equal-letter runs of the bijective Burrows-Wheeler transform. Theor. Comput. Sci. 1027: 115004 (2025).
- Christina Boucher, Davide Cenzato, Zsuzsanna Lipták, Massimiliano Rossi, Marinella Sciortino. r-indexing the eBWT. Inf. Comput. 298: 105155 (2024).
- Nicola Cotumaccio, Giovanna D’Agostino, Alberto Policriti, Nicola Prezza. Co-lexicographically Ordering Automata and Regular Languages - Part I. Journal of the ACM (JACM), 2023.
- Tomasz Kociumaka, Gonzalo Navarro, Nicola Prezza, Towards a Definitive Compressibility Measure for Repetitive Sequences, in IEEE Transactions on Information Theory (IEEE TIT), 2023.
Conference papers
- Nicola Cotumaccio, Giovanna D'Agostino, Daniel Gibney, Alberto Policriti, Nicola Prezza, Sharma V. Thankachan. Wheeler Graphs and Wheeler Languages. The Expanding World of Compressed Data 2025: 12:1-12:28.
- Davide Cenzato, Zsuzsanna Lipták, Nadia Pisanti, Giovanna Rosone, Marinella Sciortino.
BWT for String Collections. The Expanding World of Compressed Data 2025: 3:1-3:29. - Lorenzo Carfagna, Carlo Tosoni. Analysing New Entropy Measures for Tries. SPIRE 2025: 18-27.
- Ruben Becker, Nicola Cotumaccio, Sung-Hwan Kim, Nicola Prezza, Carlo Tosoni. Encoding Co-Lex Orders of Finite-State Automata in Linear Space. CPM 2025: 15:1-15:17.
- Jarno N. Alanko, Ruben Becker, Davide Cenzato, Travis Gagie, Sung-Hwan Kim, Bojana Kodric, Nicola Prezza. The Trie Measure, Revisited. CPM 2025: 19:1-19:20.
- Hideo Bannai, Philip Bille, Inge Li Gørtz, Gad M. Landau, Gonzalo Navarro, Nicola Prezza, Teresa Anna Steiner, Simon Rumle Tarnow. Text Indexing for Simple Regular Expressions. CPM 2025: 20:1-20:16.
- Ruben Becker, Giuseppa Castiglione, Giovanna D'Agostino, Alberto Policriti, Nicola Prezza, Antonio Restivo, Brian Riccardi. Universally Wheeler Languages. DLT 2025: 45-60.
- Jarno N. Alanko, Davide Cenzato, Nicola Cotumaccio, Sung-Hwan Kim, Giovanni Manzini, Nicola Prezza. Computing the LCP Array of a Labeled Graph. CPM 2024: 1:1-1:15.
- Ruben Becker, Davide Cenzato, Sung-Hwan Kim, Bojana Kodric, Riccardo Maso, Nicola Prezza. Random Wheeler Automata. CPM 2024: 5:1-5:15.
- Giovanni Manzini, Alberto Policriti, Nicola Prezza, Brian Riccardi. The Rational Construction of a Wheeler DFA. CPM 2024: 23:1-23:15.
- Ruben Becker, Matteo Canton, Davide Cenzato, Sung-Hwan Kim, Bojana Kodric, Nicola Prezza. Sketching and Streaming for Dictionary Compression. DCC 2024: 213-222.
- Ruben Becker, Sung-Hwan Kim, Nicola Prezza, Carlo Tosoni. Indexing Finite-State Automata Using Forward-Stable Partitions. SPIRE 2024: 26-40.
- Davide Cenzato, Francisco Olivares, Nicola Prezza. On Computing the Smallest Suffixient Set. SPIRE 2024: 73-87.
- Patrick Dinklage, Johannes Fischer, Nicola Prezza. Top- k Frequent Patterns in Streams and Parameterized-Space LZ Compression. SEA 2024: 9:1-9:20.
- Elena Biagi, Davide Cenzato, Zsuzsanna Lipták, Giuseppe Romana. On the Number of Equal-Letter Runs of the Bijective Burrows-Wheeler Transform. ICTCS 2023: 129-142.
- Ruben Becker, Davide Cenzato, Sung-Hwan Kim, Bojana Kodric, Alberto Policriti and Nicola Prezza. Optimal Wheeler Language Recognition. In: proceedings of SPIRE 2023.
- Nicola Cotumaccio, Travis Gagie, Dominik Koeppl and Nicola Prezza. Space-time Trade-offs for the LCP Array of Wheeler DFAs. In: proceedings of SPIRE 2023.
- Ruben Becker, Manuel Cáceres, Davide Cenzato, Sung-Hwan Kim, Bojana Kodric, Francisco Olivares, Nicola Prezza. Sorting Finite Automata via Partition Refinement. In: proceedings of ESA 2023.
- Alessio Conte, Nicola Cotumaccio, Travis Gagie, Giovanni Manzini, Nicola Prezza, Marinella Sciortino. Computing matching statistics on Wheeler DFAs. In: proceedings of DCC 2023.
- Sung-Hwan Kim, Francisco Olivares, Nicola Prezza. Faster Prefix-Sorting Algorithms for Deterministic Finite Automata. In: proceedings of CPM 2023.
Team
Current members
Nicola Prezza
Principal Investigator
Bio
Nicola received his PhD in Computer Science from the University of Udine (Italy) in 2017. He spent research periods (as a postdoc) at the universities of Pisa (Italy) and DTU (Denmark), and later has been Assistant professor at LUISS (Rome). He is now Associate professor at Ca' Foscari university of Venice. Nicola Prezza's research is focused on the interplay between data structures, data compression, and regular language theory. In 2018 he received the "best italian young researcher in Theoretical Computer Science" award from the italian chapter of the European Association for Theoretical Computer Science (IC-EATCS).
Ruben Becker
Assistant Professor, Tenure Track (RTT)
Bio
Ruben holds a PhD degree from Saarland University (Germany). While being a doctoral student (2014-2018) he was a member of the Algorithms and Complexity department at Max Planck Institute for Informatics. He was then a Postdoctoral researcher in the Computer Science department of the Gran Sasso Science Institute in L'Aquila, Italy (2018-2022). Since November 2022 he is an assistant professor (RTD-A) at Ca' Foscari University of Venice. Ruben's research revolves around the topics algorithms, graphs, and randomness. For his PhD thesis, he obtained the Dr. Eduard Martin Preis that is awarded yearly to the best dissertation within the Faculty of Computer Science and Mathematics at Saarland University.
Alessio Campanelli
PhD student
Bio
Alessio graduated in October 2024 in Computer Science at Ca' Foscari University of Venice (Italy), with a thesis on Bioinformatics. He then started his PhD programme at the same university, with the REGINDEX research group. He is a competitive programming enthusiast and is main area of interest is compact data structures.
Davide Cenzato
Postdoc
Bio
After graduating in Medical bioinformatics in 2019, Davide received his Ph.D. in Computer Science at the University of Verona (Verona, Italy). He then joined Ca' Foscari University of Venice (Venice, Italy) in February 2023 as a postdoc researcher in the REGINDEX group. His main research interests are algorithms and data structures on strings and graphs; in particular, he focuses on algorithms for processing and indexing genomic string collections and pangenomes, with a special emphasis on providing efficient implementations.
Riccardo Maso
PhD student
Bio
Riccardo graduated in Computer Science from Ca' Foscari University of Venice (Italy) in March 2025. He later joined the REGINDEX research group at the same university as a pre-doctoral research fellow, and in September 2025 he began his PhD.
Daniel Puttini
PhD student
Bio
Daniel earned his master's degree in Computer Science from the University of Parma in March 2023, having completed a thesis in bioinformatics. Following graduation, he collaborated with the Department of Food and Drugs at the University of Parma for six months. In November 2023, he joined the REGINDEX research group at Ca' Foscari University of Venice (Venice, Italy), where he is currently engaged as a (“pre-doc”) research fellow, eagerly anticipating the commencement of his doctoral studies.
Davide Tonetto
PhD student
Bio
Davide earned his master’s degree in Computer Science and Information Technology from Ca’ Foscari University of Venice in October 2025, where he completed a thesis on the compression of finite languages. He has since joined the REGINDEX research group at Ca’ Foscari University as a doctoral student, continuing his academic journey in computer science and pursuing his passion for study and research.
Carlo Tosoni
PhD student
Bio
After graduating in 2021 with a degree in computer engineering from the University of Perugia, Carlo enrolled in the master's degree programme in computer science at the University of Pisa, where he graduated in July 2023. His chosen curriculum, 'Big Data Technologies', aimed to develop the skills needed to manage large collections of data in the most efficient possible manner. Two months later, he started a PhD programme at Ca' Foscari University in Venice, in the research group of Nicola Prezza.
Past members
Bio
Sung-Hwan received his PhD degree in computer science and engineering from Pusan National University, Busan, South Korea, in 2018. Since then, he had been a postdoctoral researcher at Pusan National University until he joined Nicola Prezza's team in September 2022. He has been interested in several research topics including metric indexing, string matching, and sequence alignment, among which the most favorite is design and analysis of simple algorithms and space-efficient data structures for pattern matching problems.
Bojana Kodric
Postdoc
Bio
After studying Mathematics at the University of Zagreb (Zagreb, Croatia), Bojana completed a PhD in Theoretical Computer Science at the Max Planck Institute for Informatics (Saarbrücken, Germany) in 2018. Subsequently, she was a PostDoc at the Gran Sasso Science Institute (L’Aquila, Italy), until she joined Ca’ Foscari University of Venice (Venice, Italy) as a PostDoc on the REGINDEX project. Her areas of interest are Algorithms and Complexity and Algorithmic Game Theory.